Low-level API
Contents:
Low level array-based operations are used to implement the genomic interval operations on dataframes.
import itertools
import numpy as np
import matplotlib
import matplotlib.pyplot as plt
import pandas as pd
import bioframe as bf
import bioframe.vis
from bioframe.core import arrops
starts1, ends1 = np.array([
[1,5],
[3,8],
[8,10],
[12,14]
]).T
starts2, ends2 = np.array([
[4,8],
[10,11],
]).T
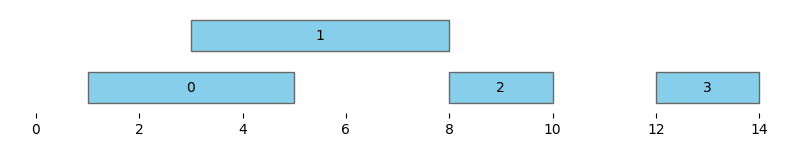
bf.vis.plot_intervals_arr(
starts = starts1,
ends = ends1,
xlim = (-0.5,14.5),
labels = np.arange(0,starts1.shape[0]),
show_coords = True)
bf.vis.plot_intervals_arr(
starts = starts2,
ends = ends2,
colors = 'lightpink',
xlim = (-0.5,14.5),
labels = np.arange(0,starts2.shape[0]),
show_coords = True)

arrops.overlap_intervals(starts1, ends1, starts2, ends2)
array([[0, 0],
[1, 0]])
arrops.overlap_intervals_outer(starts1, ends1, starts2, ends2)
(array([[0, 0],
[1, 0]]),
array([2, 3]),
array([1]))
arrops.merge_intervals(starts1, ends1, min_dist=0)
(array([0, 0, 0, 1]), array([ 1, 12]), array([10, 14]))
arrops.merge_intervals(starts1, ends1, min_dist=None)
(array([0, 0, 1, 2]), array([ 1, 8, 12]), array([ 8, 10, 14]))
arrops.merge_intervals(starts1, ends1, min_dist=2)
(array([0, 0, 0, 0]), array([1]), array([14]))
arrops.complement_intervals(starts1, ends1)
(array([ 0, 10, 14]),
array([ 1, 12, 9223372036854775807]))